What we do


The trillions of microorganisms living in our gut (as well as the 70 gigatons carbon of microbes on the planet) perform a complex ballet of metabolic interactions between each other and their environment, giving rise to a myriad of biotransformations.
We study the flexibility of microorganisms in adapting their nutrient needs and their interactions with surrounding microbes in varying environments with a focus on how this may impact health and disease in the human gut.
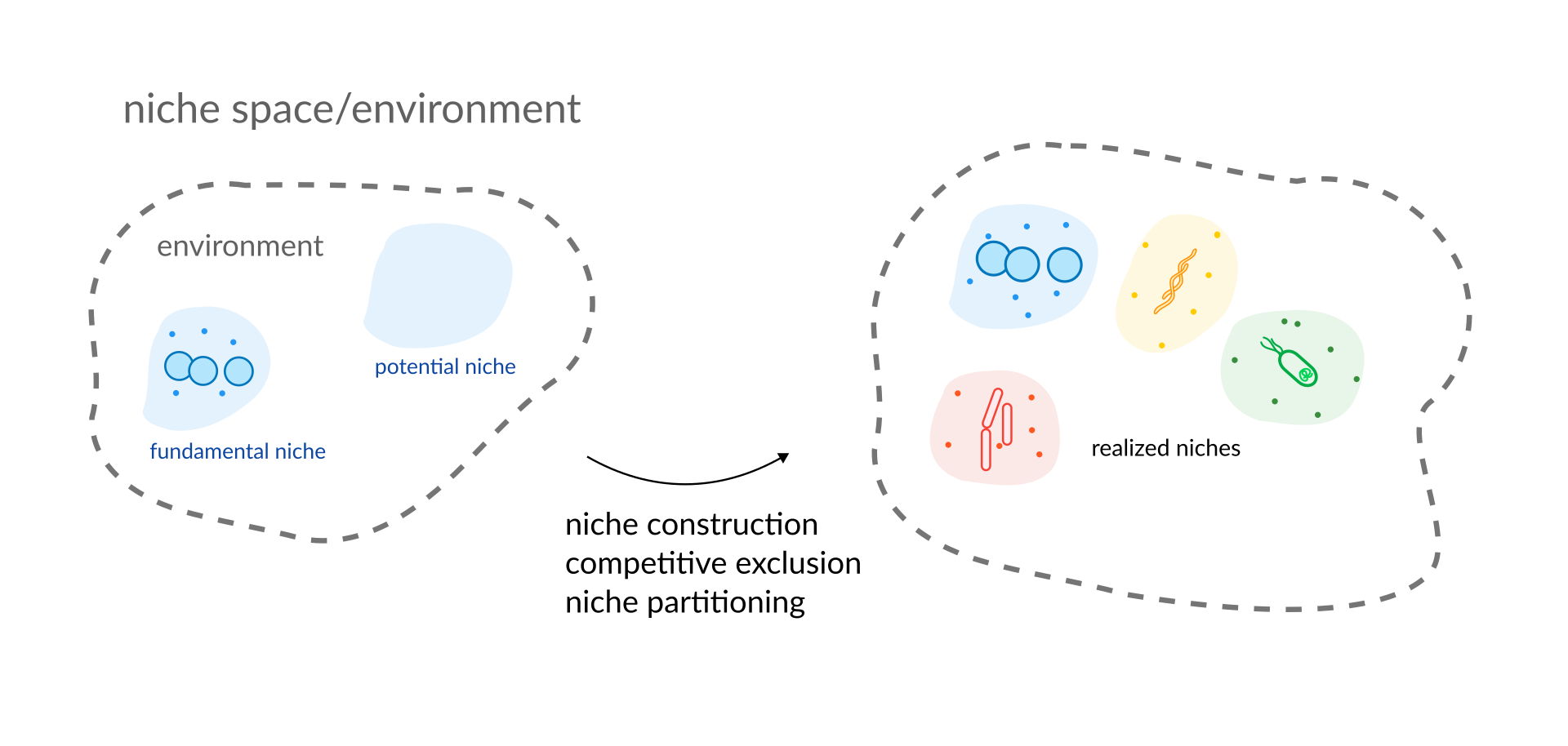
Questions we are interested in are:
We address these questions using computational and wetlab approaches including metagenomics, metabolomics, modeling, and single-cell culturomics.
A large fraction of anything we eat will eventually make its way into the intestine where it comes into contact with the trillions of microorganisms residing there. Those commensal microbes have a staggering flexibility in consuming or modifying compounds in their environment affecting the chemical profile of the human intestine.
We are interested in the role microbes play in determining the chemical make-up of the human gut. This is not limited to dietary compounds but also pharmaceutical drugs (especially statins), host-derived compounds (such as hepatic metabolites and mucins), and xenobiotics.
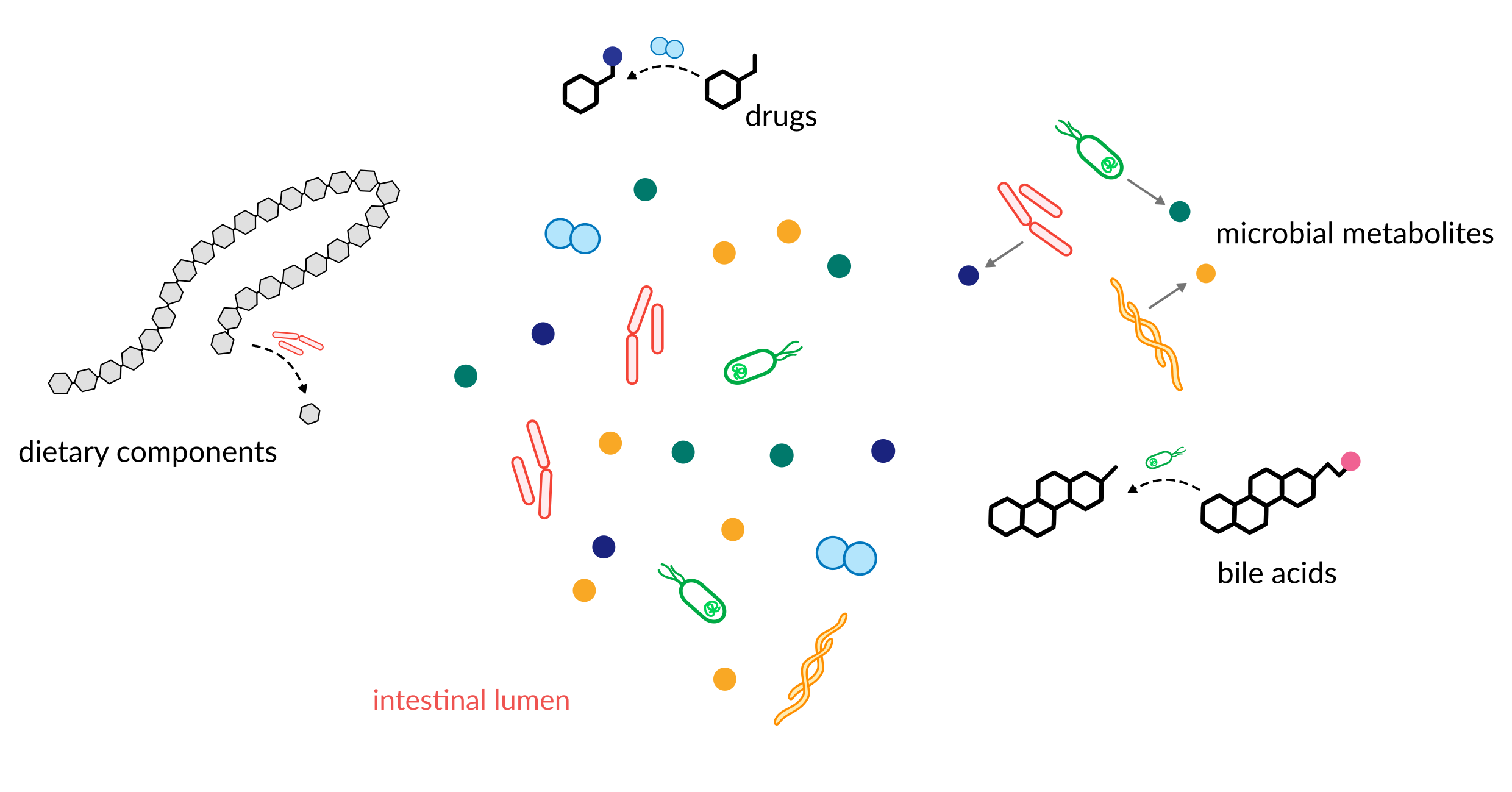
Questions we are interested in are:
In the last decades we have generated a panacea of information on the gut microbiome. We have learned that the gut microbiome is altered in different diseases and that the transfer of the microbiome from one individual to another can alleviate some diseases. However, to leverage microbiome treatments in the clinic we need clear and defined strategies for interventions and their effects on the human host.
We leverage combinations of computational strategies and simplified experimental models to study and predict the effect of interventions on the human gut microbiome and their impact on the host.
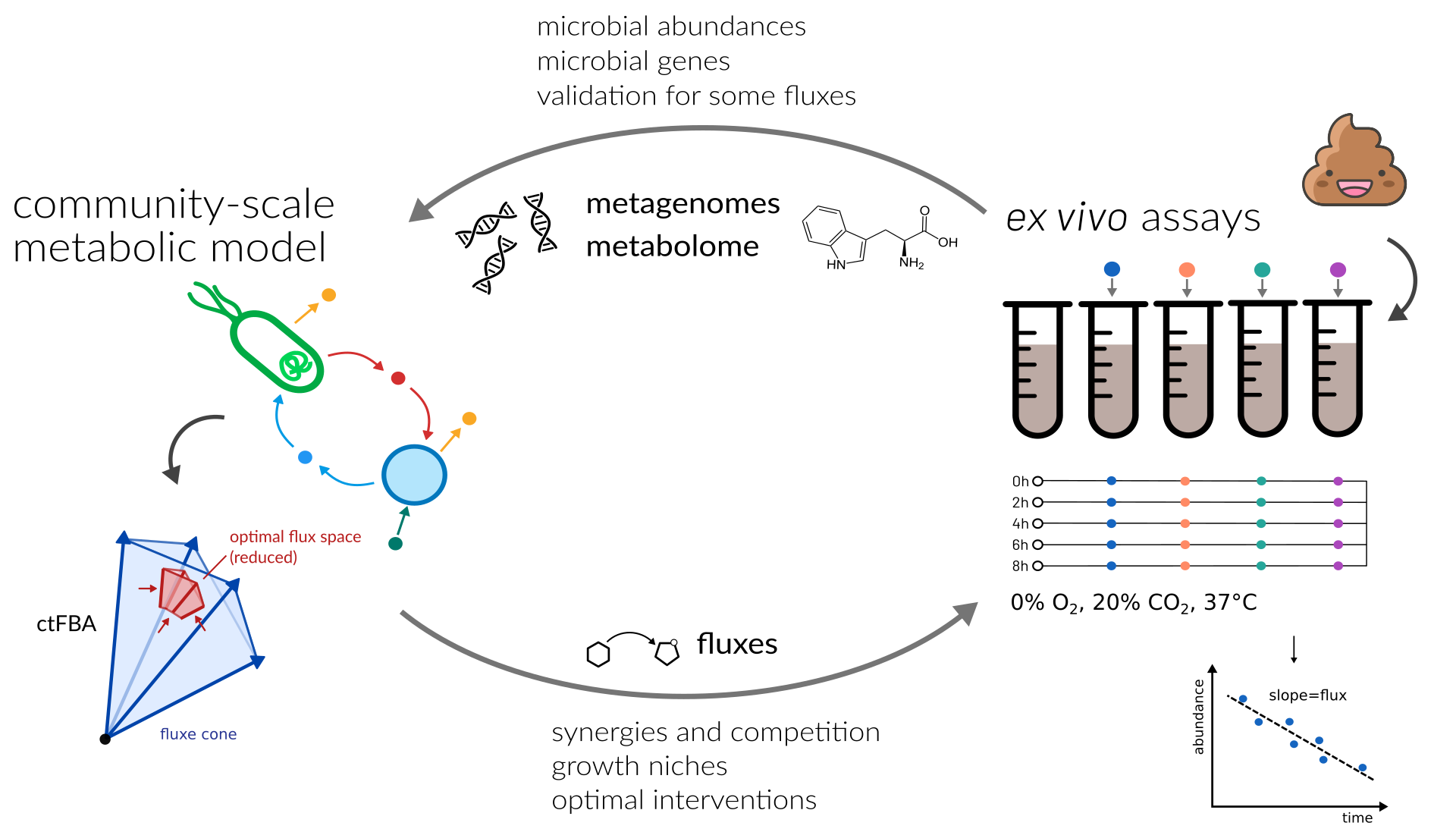
Questions we are interested in are: